Welcome to PyRaDiSe’s Documentation#
PyRaDiSe (Python package for Radiotherapy-oriented and DICOM-based auto-Segmentation) is an open-source Python package for building clinically applicable radiotherapy-oriented auto-segmentation solutions. This package addresses two main challenges of auto-segmentation in clinical radiotherapy: data handling and conversion from and to DICOM-RTSS. Besides a radiotherapy-oriented data model and conversion capabilities, PyRaDiSe provides a profound set of extensible processing filters for fast prototyping and development of clinically deployable auto-segmentation pipelines. Therefore, PyRaDiSe is a highly flexible and extensible toolbox, allowing for narrowing the gap between data science and clinical radiotherapy research and speeding up development cycles.
Main Features#
The main features of PyRaDiSe are data handling, conversion from and to DICOM-RTSS, and data processing,
including deep learning model inference. The intended use of PyRaDiSe in the radiotherapy environment is depicted in
Fig. 8. The DICOM and other discrete medical image file formats, such as NIFTI, are imported
into the provided data model using the fileio package. In contrast to the standard way of loading
DICOM data, this package provides comprehensive and flexible import routines that consider data relation details and
automate import steps, such as registering DICOM images if DICOM registration files are available. However, in some
cases, the DICOM standard does not provide sufficient information for automation, requiring minimal human interaction
for resolution. In addition, discrete medical images also suffer from the lack of identification data needed for
automation. However, the fileio package offers the necessary methods to address these issues with
flexible approaches and prototypes. Furthermore, the fileio package provides routines to select
specific entities from the available data before loading by generating filterable pre-loading information
(so-called SeriesInfo) so that the computation time and memory usage
for loading is minimal. Finally, after the data is loaded, it is represented using the data model implemented in the
data package. All downstream tasks are performed using the simple and extensible radiotherapy-oriented
data model from this step on.
After loading, the data is either converted and written to a file or processed using routines from the
process package. This package includes functionality and prototypes for pre-processing, deep
learning model inference, and post-processing with a similar mode of operations as well-known medical image libraries,
such as SimpleITK or ITK. However, in contrast to other libraries, the process package offers a
mechanism for guaranteeing reproducibility and limited invertibility.
After processing or loading, the altered data can be written to disk using a versatile writer from the
fileio package to save the data as either a discrete image file or as DICOM-RTSS. In addition, some of
the offered writers provide the additional functionality to copy the existing data from the source to the
target directory. This feature is handy if the developed auto-segmentation solution will be deployed to the clinical
environment or the cloud, where the original input data should remain unmodified.
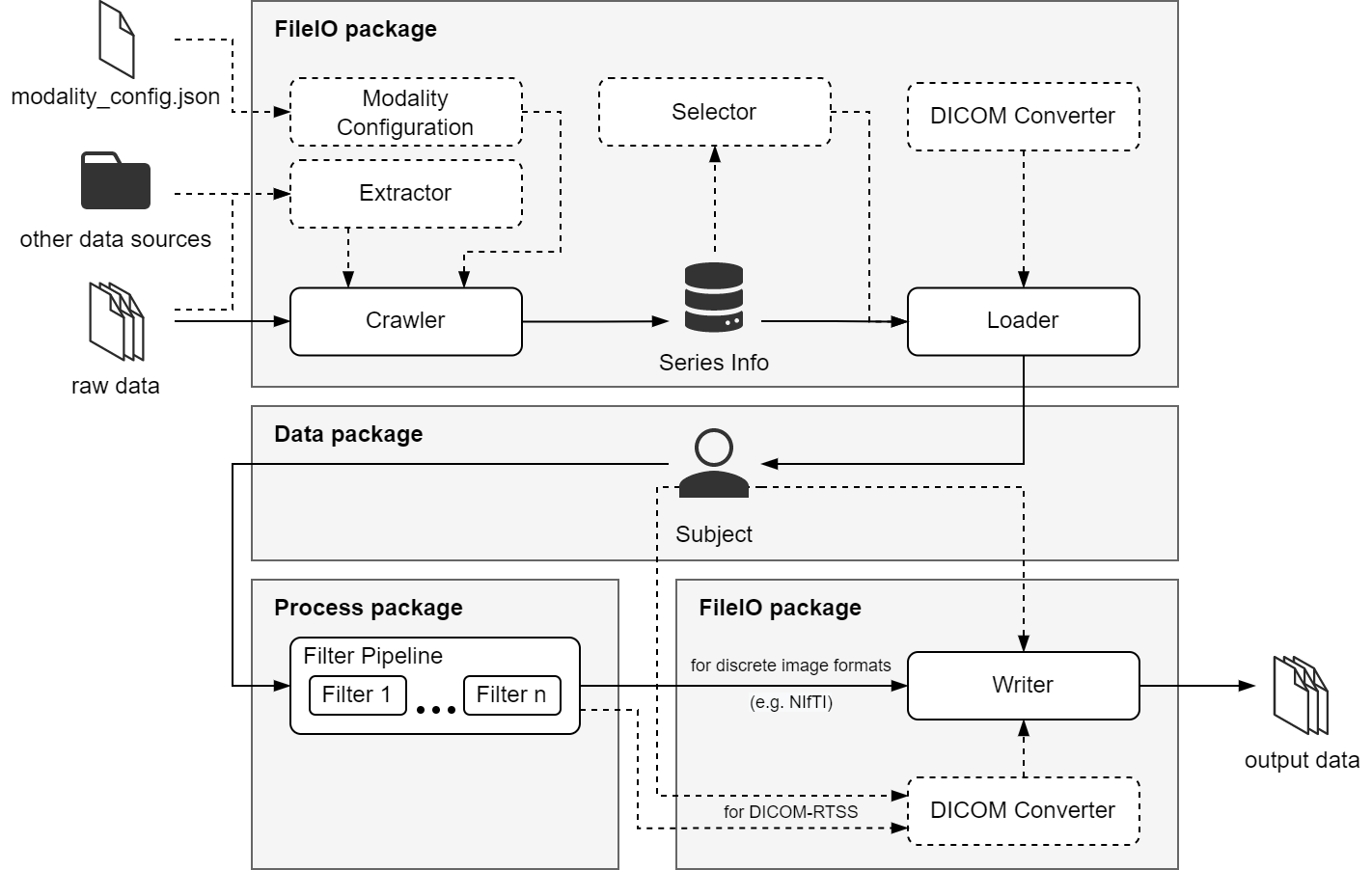
Fig. 8 Schematic illustration of PyRaDiSe in the radiotherapy environment#
Getting Started#
If you are new to PyRaDiSe we recommend to start with the Installation and Examples pages to get used to the package. Furthermore, the API Reference page provides an overview of the available modules and classes.
If you have used PyRaDiSe before, please consider the Change History page for the latest changes. We also want to encourage you to make pull requests to the PyRaDiSe repository on GitHub if you have any improvements or suggestions. Furthermore, we would be happy to hear from you if you have any questions or comments.
Finally, the Acknowledgments page provides an overview of the acknowledgments for the development of PyRaDiSe.
Citation#
If you use PyRaDiSe in your research, please acknowledge it by citing our paper:
BibTeX entry:
@article{Ruefenacht2023,
author = {Rüfenacht, Elias and Kamath, Amith and Suter, Yannick and Poel, Robert and Ermis, Ekin and Scheib, Stefan and Reyes, Mauricio},
title = {{PyRaDiSe: A Python package for DICOM-RT-based auto-segmentation pipeline construction and DICOM-RT data conversion}},
journal = {Computer Methods and Programs in Biomedicine},
doi = {10.1016/j.cmpb.2023.107374},
issn = {0169-2607},
year = {2023}
}